Vascular & Developmental Biology
Explore the vascular and developmental biology research programs and labs below and visit our Center for Vascular & Developmental Biology page to learn more.
Paul Burridge, PhDInvestigating the pharmacogenomics of chemotherapy-induced toxicity and electrophysiological disorders using human induced pluripotent stem cells.
Investigating the pharmacogenomics of chemotherapy-induced toxicity and electrophysiological disorders using human induced pluripotent stem cells.
The Burridge Lab works in the fields of pharmacogenomics (precision medicine), cardio-oncology, cardiovascular disease modeling, regenerative medicine, cancer, and cultivated meat, using induced pluripotent stem cells (iPSCs) as our model.
We specialize in large-scale hiPSC projects that required reprogramming, sequencing, editing, differentiating, and phenotyping hundreds of hiPSC lines.
Ongoing projects include:
- Studying the role of the genome in complex disease traits, understanding how single genetic variants modulate drug efficacy and toxicity, predicting which patients will experience successful therapy or suffer adverse drug responses, and discovering new drugs to overcome these complications.
- Developing new tools to reprogram, culture, and differentiate iPSC in an efficient and cost-effective manner to improve the use, scale, and ubiquity of these across all areas of research.
- Studying the genomics of arrhythmia and sudden cardiac death and devising precision medicine-led drug discovery.
- The applications of iPSC and direct reprogramming in regenerative medicine, repairing the heart after a heart attack, or ameliorating the effects of heart failure.
- How heritable (germline) variation influences cancer progression, oncology drug efficacy, and acquisition of resistance.
- The applications of iPSC-derived cells for cultivated meat.
Contact Us
Searle 8-450 320 E. Superior St.Chicago, IL 60611United States
paul.burridge@northwestern.edu
See the faculty profile for Paul W. Burridge, PhD, or visit Paul Burridge Lab website to learn more.
Matthew Feinstein, MDConducting translational research on immune and inflammatory triggers of cardiovascular diseases.
Karen Ho, MDSeeking a better understanding of the mechanisms of atherosclerosis and neointimal hyperplasia as a cause of restenosis after vascular surgery in order to prevent and treat these diseases.
We are specifically interested in how the gut microbiome and microbe-derived metabolites regulate atherosclerosis and arterial remodeling after vascular surgery. We use microbiome manipulations, germ-free mouse models, surgical models of arterial injury, genetic and diet-induced models of atherosclerosis, cell/molecular biology techniques, targeted metabolomics and cultivation-independent analysis of microbial communities to obtain a phenomenological and mechanistic understanding these diseases, which will translate to the clinical realm.
See the faculty profile for Karen J. Ho, MD, or visit the Ho Lab site to learn more.
Philip Iannaccone, MD, PhDStudying how the Sonic hedgehog signal in vertebrates is mediated by three C2H2 zinc finger transcription factors, GLI1, GLI2 and GLI3.
Our lab studies how the Sonic hedgehog signal in vertebrates is mediated by three C2H2 zinc finger transcription factors, GLI1, GLI2 and GLI3. We have shown that near identity of gene sequence exists between mouse and human GLI1 and that GLI1 functions as a transcription factor and that a critical domain in the COOH end of the protein is a transactivator with VP16-like structure.
We established that:
- GLI1 protein regulates a set of genes that coordinately control proliferation and may in part explain malignant transformation by misexpression of GLI1
- Non-canonical activation of GLI1 by c-myc short-circuiting patched and smoothened trans-membrane proteins in lymphoma
- GLI1 binds TAF9 at the gene target and competes with P53 for this adapter protein providing an explanation for GLI1 oncogenic activity
We are working out the mechanisms of autoregulation of GLI1 by molecular analysis of cis and trans effectors using gene editing techniques. We have established a strong intronic enhancer region that in part explains GLI autoregulation. We are establishing key protein-protein interactions during assembly of the transcription machinery. We have developed chimeras and a method to identify cell lineage in visceral organs. We established that chemically induced tumors and preneoplastic lesions are clonal, the first lab to do this. The patterns in normal tissue derive from the allocation of stem cells during development. We show that the patterns are fractal, likely arise from iterating simple division rules and do not require cell movement for complex, highly variegated patterns to form.
In the cornea epithelial cells are assorted in spirals. Absence of the pattern is associated with Pax6 mutations and keratopathies. Using finite element models we have shown the patterns correlate with path lines of maximum stress/strain, suggesting that shearing forces are responsible for the pattern.
See the faculty profile for Philip Iannaccone, MD, PhD to learn more.
Luisa Iruela-Arispe, PhDExploring molecular regulation of angiogenesis and vascular homeostasis.
Currently, the laboratory is investigating the mechanisms behind the formation of vascular tumors and vascular anomalies through the identification of critical regulatory nodes that maintain vascular homeostasis and control endothelial proliferation in the context of flow.
Mechanotransduction in endothelial and smooth muscle cells and heterotypic endothelial – macrophage cell interactions remain areas of interest, along with VEGF and Notch signaling. An additional focus of the lab is to understand vascular resilience and dissect the molecular mechanisms behind response to stressors, including aging.
See the faculty profile for Luisa Iruela-Arispe, PhD, or visit the Arispe Lab website to learn more.
Yuzhi Jia, MD, PhDStudying the mechanisms that regulate the transcriptional activities of nuclear receptors.
Nuclear receptors constitute a large superfamily of transcription factors that regulate a diverse array of biological processes, including development and neoplasia, as well as energy and xenobiotic metabolism. In order to initiate transcription, some more factors are required to interact with nuclear receptors and to regulate their activities. These factors are collectively called "nuclear receptor co-factors." Several co-factors have been identified by our studies. Generating mouse models using gene targeting technology to investigate the functions of these genes is in process.
See the faculty profile for Yuzhi Jia, MD, PhD, to learn more.
Jing Jin, MD, PhDSeeking to understand the molecular mechanisms of kidney and vasculature diseases.
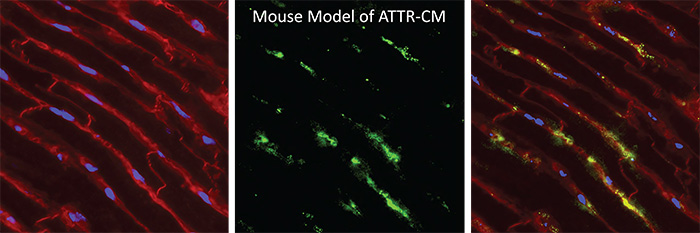
The Jin Lab is interested in understanding the molecular mechanisms of kidney and vasculature diseases. Cell junction and matrix proteins play a major role in the disease etiology and progression. We study how vascular and glomerular basement membrane (GBM) matrix proteins are interwoven, and the mechanisms for physiological and pathological GBM remodeling during repair. Specifically, we use mass spectrometry to map the patterns of post-translational modifications such as hydroxylation and glycosylation on the GBM collagen and study how these affect the meshwork topology. Ultimately we hope such knowledge may help to devise targeted therapies for a broad range of kidney and vascular diseases
The lab is generally interested in the pathological mechanisms of kidney and vascular diseases. We take a proteomic approach to study molecules that serve structural or functional roles in kidney filtration. Particularly, we are trying to understand how the kidney podocytes maintain and regulate their slit diaphragm, as well as their interactions with the glomerular basement membrane.
See the faculty profile for Jing Jin, MD, PhD, to learn more.
Evangelos Kiskinis, PhDAddressing fundamental aspects of the biology of human neurons in the context of physiological conditions and in the context of disease.
We have two clinical interests: motor neuron disease and epileptic channelopathies. We utilize patient-specific induced pluripotent stem cells and direct reprogramming methods to generate different neuronal subtypes implicated in these diseases. We then study these cells by a combination of genomic approaches and functional physiological assays. Our hope is that these disease model systems will help us identify points of effective and targeted therapeutic intervention.
See the faculty profile for Evangelos Kiskinis, PhD, or visit the Kiskinis Lab website to learn more.
Tsutomu Kume, PhDFocused on cardiovascular development and disease with an emphasis on the formation of the vascular system that is composed of blood and lymphatic vessels; we study the cellular and molecular mechanisms that control neovascularization in physiological and pathological processes using various mouse models.
Focused on cardiovascular development and disease with an emphasis on the formation of the vascular system that is composed of blood and lymphatic vessels; we study the cellular and molecular mechanisms that control neovascularization in physiological and pathological processes using various mouse models.
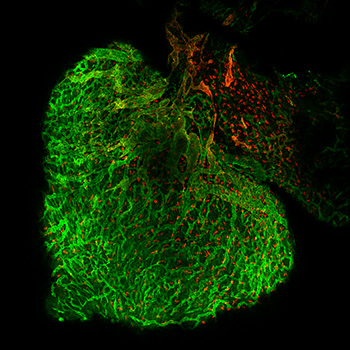
The Kume Laboratory is recognized for studies highlighting the role of a family of transcription factors termed Forkhead/Fox in diverse biological processes. Key observations include studies elucidating the role of Foxc1 and Foxc2 as (1) factors that serve as essential regulators of cardiovascular development, (2) key factors of angiogenesis and lymphangiogenesis, and (3) important regulators of the anterior eye segment. These research efforts have spawned novel areas of investigation with therapeutic implications for numerous biological systems, including the biology of the cardiovascular systems.
Studies in the lab have recently extended toward the area of the organ-specific vasculature, and we have established a research team to investigate mechanisms of tissue repair and regeneration using a variety of mouse models including ischemia-reperfusion injury.
See the faculty profile for Tsutomu Kume, PhD, to learn more. William A. Muller, MD, PhDStudying the inflammatory response at the cellular and molecular level.
We are focused on the process of diapedesis, the "point of no return" in inflammation where leukocytes squeeze between tightly apposed endothelial cells to enter the site of inflammation. We have identified and cloned several molecules that are critical to the process of diapedesis (PECAM (CD31), CD99 and VE-cadherin) and are studying how they regulate the inflammatory response using in vitro and in vivo models.
We have recently described the Lateral Border Recycling Compartment (LBRC), a novel para-junctional organelle that contains PECAM and CD99 and is critical for diapedesis to occur, seemingly independent of the leukocyte type, inflammatory stimulus, or vascular bed. We are currently investigating how this compartment regulates diapedesis in the hope of finding novel targets for anti-inflammatory therapy. Our discovery that a single splice form of a single kinesin light chain is required for moving the LBRC to the site of diapedesis has led to the design of a prototype anti-inflammatory drug that is currently being tested in several mouse models.
Our inflammatory models include atherosclerosis, myocardial infarction, ischemia/reperfusion injury, stroke, dermatitis, multiple sclerosis, peritonitis, and rheumatoid arthritis. We are also using 4-dimensional intravital microscopy to view the inflammatory response in real time in living animals.
Visit the faculty profile of William A. Muller, MD, PhD, to learn more.
Guillermo Oliver, PhDCharacterizing the functional roles of the lymphatic vasculature in health and disease; we are particularly interested in the role of lymphoangiocrine factors and lymphatic metabolism in regulating organogenesis and cardiac repair using mouse models.
Characterizing the functional roles of the lymphatic vasculature in health and disease; we are particularly interested in the role of lymphoangiocrine factors and lymphatic metabolism in regulating organogenesis and cardiac repair using mouse models.
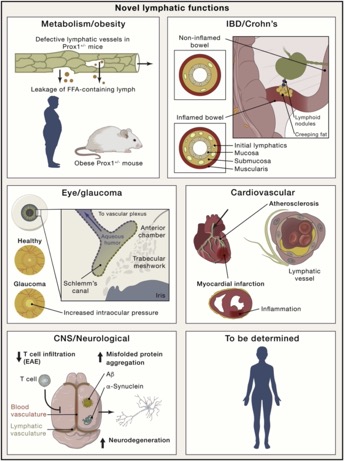
Morphological or functional defects in the lymphatic vasculature have now been uncovered in several pathological conditions such as obesity, cardiovascular disease, inflammation, glaucoma and neurological disorders such as Alzheimer’s disease (Oliver et al., Cell 2020). We identified the transcription factor Prox1 as the first specific marker of lymphatic endothelial cells (LECs) (Wigle and Oliver, Cell 1999). We also demonstrated that Prox1 is necessary for LEC specification (Wigle et al., EMBO J 2002) and that LEC fate is plastic and reprogrammable (Johnson et al., Genes & Dev 2008). Years ago, we reported that obesity can be consequence of lymphatic vasculature defects (Harvey et al., Nature Genetics 2005). We have characterized the early steps of lymphatic vasculature formation and showed that a Prox1-Vegfr3 feedback loop is required to regulate the number of LEC progenitors and to maintain their identity (Srinivasan et al., Genes & Dev 2014). Recently we determined that mitochondrial controls the Prox1-Vegfr3 feedback loop during LEC fate specification (Ma et al., Science Advances 2021). We also identified a novel functional role of lymphatics during cardiac growth and repair (Liu et al., Nature 2020).
See the faculty profile for Guillermo Oliver, PhD, or visit the Oliver Lab website to learn more.
Susan E. Quaggin, MDUnderstanding the role of the ocular endothelium in vision with the aim of discovering therapeutic targets to be translated into patient care.
Endothelial dysfunction is a major cause of vision loss, playing a key role in diseases including age-related macular degeneration, diabetic retinopathy and glaucoma; we focus on understanding the role of the ocular endothelium in vision with the aim of discovering therapeutic targets to be translated into patient care.
See the faculty profile for Susan Quaggin, MD, to learn more.
Jeffrey N. Savas, PhDDetermining how protein mishandling contributes to synaptic dysfunction and aging.
Our work is centered on elucidating the pathogenic mechanisms that drive synaptopathies and proteinopathies in the mammalian nervous system. As a first step, we use reductionist biochemical approaches with discovery-based proteomic analysis to generate unbiased hypothesizes. Next, we test and characterize candidate proteins and pathways for potential functional relevance with molecular approaches. Ultimately, the goal is to identify keystone proteins and proteomes as potential therapeutic targets to hopefully ameliorate neurodevelopmental and degenerative diseases.
See the faculty profile of Jeffery N. Savas, PhD, or visit the Savas Lab website to learn more.
Beatriz Sosa-Pineda, PhDUsing mouse models and cutting-edge techniques to study pathological conditions that increase injury during pancreatitis; we also use mouse models and in vitro approaches to study mechanisms that promote pancreatic tumor aggressiveness.
Using mouse models and cutting-edge techniques to study pathological conditions that increase injury during pancreatitis; we also use mouse models and in vitro approaches to study mechanisms that promote pancreatic tumor aggressiveness.
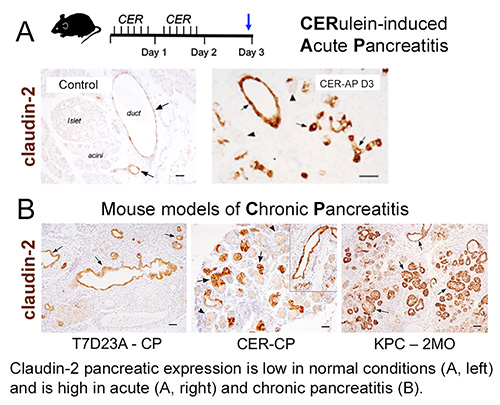 Pancreatitis is an inflammatory disease triggered by environmental and genetic factors. Understanding how individual risk factors influence the pathologic outcome in pancreatitis should help the management and diagnosis of the disease. Genetic studies found that mutations in the gene CLDN2 (encoding the tight junction protein claudin-2) sensitize patients to alcoholic chronic pancreatitis. The functional role of claudin-2 in pancreas homeostasis and pancreatitis has not been investigated and this is a major focus of our current research.
Pancreatitis is an inflammatory disease triggered by environmental and genetic factors. Understanding how individual risk factors influence the pathologic outcome in pancreatitis should help the management and diagnosis of the disease. Genetic studies found that mutations in the gene CLDN2 (encoding the tight junction protein claudin-2) sensitize patients to alcoholic chronic pancreatitis. The functional role of claudin-2 in pancreas homeostasis and pancreatitis has not been investigated and this is a major focus of our current research.
Another project in the lab with relevance to the mission of FCVRRI investigates the role of the pancreatic lymphatic vasculature in homeostasis and in pancreatitis.
Pancreatic cancer has one of the worst cancer survival rates worldwide. Late detection of the disease is frequent and at stages when the tumor already metastasized to vital organs (e.g., the liver and lungs). Understanding the underlying biology of pancreatic tumors should help developing new diagnostic methods and better therapies. Our laboratory uses mouse models and human pancreatic cancer cell lines to uncover new regulatory mechanisms that confer pancreatic tumor aggressiveness.
See the faculty profile for Beatriz Sosa-Pineda, PhD, or visit the Sosa-Pineda Lab site to learn more.
Ed Thorp, PhDInvestigating the cell and molecular biology of the cardiovascular system in steady state versus during inflammation.
Investigating the cell and molecular biology of the cardiovascular system in steady state versus during inflammation.
See the faculty profile for Ed Thorp, PhD, or visit the Thorp Lab website to learn more.
Lisa D. Wilsbacher, MD, PhDStudying the roles of G protein-coupled receptors in heart development and in the setting of adult heart disease.
Studying the roles of G protein-coupled receptors in heart development and in the setting of adult heart disease.
The goal of our research is to define key signaling pathways in cardiac development and cardiomyocyte maintenance in the setting of pathological stress. Currently, the laboratory investigates the G protein-coupled receptor sphingosine-1-phosphate receptor 1 (S1P1) and its unexpected role in cardiomyocyte proliferation and cardiac development. We aim to identify the signaling mechanisms that underlie these cardiac developmental effects and to investigate whether S1P1 signaling contributes to cardiac remodeling in the adult heart.
See the faculty profile for Lisa Wilsbacher, MD, PhD, or visit the Wilsbacher Lab site to learn more.